
A 10-year Census of the Sea undertaken by more than 2,700 scientists in 83 countries is churning up new questions about biodiversity and the role of marine creatures in the ecosystem.
The recently completed census was announced Oct. 4 in London, where Ann Bucklin, professor and head of marine sciences in the College of Liberal Arts and Sciences, represented the zooplankton section, which she led.
It was one of 14 parts of the census. Her postdoctoral researcher, Leo Blanco-Bercial, was project manager for the USA office of the global zooplankton project.
The census, which focused on species recognition, provides a baseline of information, enabling scientists to explore further at the crossroads of ecology and evolution, says Bucklin.
It also yielded information about species that will help scientists assess how marine life adapts to climate change. And it adds to our knowledge, still sparse, about the deep ocean.
The zooplankton census found more than 100 new species, adding to the 7,000 already known. While the number added might appear small – like the tiny zooplankton, which float on the ocean currents and provide a crucial link in the food chain – it is big considering how long taxonomy takes, Bucklin says.
Taxonomy – the description and cataloging of new species, their range and distribution – can be excruciatingly slow and prone to error because it is so difficult. That has curbed progress in biodiversity research, she says.
But technological advances from the sea census – such as DNA barcoding of zooplankton, Bucklin’s specialty – will speed up the process of species identification. That will likely lead to new insights into who lives where and why, and why biodiversity matters.
“Now we can ask questions about why – that’s really exciting,” Bucklin says.

The zooplankton group expects that 14,000 species of zooplankton exist, double the number now known.
Among the zooplankton group’s findings was a biodiversity “hot spot” in the deep sea off Southeast Asia, where many new zooplankton species were found.
Many of the new zooplankton species found in the census are rare, leading to questions about what they are doing and what their importance is in the ecosystem, Bucklin says.
DNA barcoding, which she likens to a “Rosetta stone” for zooplankton biodiversity, allows scientists to identify species of zooplankton based on a short DNA sequence common only to that species. It is usually found in a mitochondrial gene common to all.
Even if only a piece of an organism is caught in a net, it can be identified based on the sequence, or barcode.
Nearly a third of the 7,000 or so named zooplankton species will be barcoded by the end of the year, and that information will also allow scientists to identify the species’ closest relatives.
“Barcodes with names on them are an extremely valuable commodity,” says Bucklin.
The zooplankton part of the census, known as CMarZ, or Census of Marine Zooplankton, is working toward a “gold standard” bar code library, in which bar codes are associated with a species and genus name given by a taxonomy expert. That will make it possible to avoid the mistakes that are common in species identification.
Bucklin’s next step will be to do “deep sequencing” in collaboration with the Center for Applied Genomics Technology in the College of Liberal Arts and Sciences. The center’s sophisticated instrumentation makes it possible to take a bulk sample drawn from the ocean – “environmental sequencing” – and find as many as 100,000 DNA sequences at once.
This will speed the process of finding out which marine animals are located where, and of learning more about what their role is in a particular environment.
Ultimately the information will be important for fisheries management, to measure the health of ecosystems, and to see how species are moving north and south due to climate change, Bucklin says.
The Census of the Sea began 10 years ago, funded by the Sloan Foundation. But the zooplankton section began only in 2004, once the project’s directors were convinced that the zooplankton, while small and less “charismatic” than the big marine animals, should also be included.
The foundation’s support made possible true international collaboration, unrestricted by the borders often limiting national funding agencies, Bucklin says. The zooplankton section, whose steering committee had 23 members from 14 countries, surveyed every ocean. They met face to face every one to two years, and laboratory partners from Japan, Germany, China, and India visited Bucklin’s lab during the census.
“Real meetings – not teleconferencing – are extremely valuable for building successful scientific collaborations,” says Bucklin.
Results were announced Oct. 4 at a London press conference, where Bucklin spoke about the technological advance of DNA barcoding.
She is quoted in the National Geographic’s story on the census in its October issue: “The census sets a standard for the type of information needed now to understand what we’re doing to Earth.”
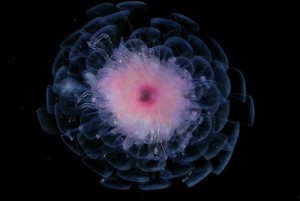



See also slides of some of the colorful and exotic zooplankton; visit the Census of the Sea video gallery; and the image gallery.
Related stories:
Marine Life Census Reveals Previously Unknown Sea Creatures
Zooplankton Expert is New Head of Marine Sciences Department


