UConn Health’s Leslie M. Loew, professor of cell biology and computer science and engineering, has received $1.3 million from the National Institutes of Health to maintain and improve a project that has been aiding international cell biology research for 21 years.
Loew is the director of the Center for Cell Analysis and Modelling at UConn Health which runs VCell. VCell, short for Virtual Cell, is an online program that allows researchers to model the behavior of any type of cell to see how it would respond in an experiment or to better analyze experimental data.
“The basic idea is that all the stuff that’s going on inside a cell should be understandable from the laws of chemistry and physics,” Loew says. “However, there’s so much going on, it’s so complex, how do you even begin to understand it?”
Loew and his collaborators answered that question in 1997 when they created VCell. The team wanted to develop a tool to help them understand the complex cellular problems they were encountering in their own research.
“We had mathematicians and computer scientists working with us. We put our heads together and realized we needed a computer simulation to solve our specific problems,” Loew says. “It started with our own research and then we realized it could be used more generally.”
VCell now has 22,000 registered users from all over the world primarily in the cell biology and biophysics fields. The new NIH grant provides funding for the next three years of the program; VCell has been continuously funded by NIH since 1998.
While the VCell tool and database are housed at UConn Health, researchers can access the program anywhere.
“The modelling can be done on a local computer, but if a researcher needs more horse power, they can use the hardware at UConn Health,” Loew says. “They can also store their models and simulation results on our database.”
VCell has two major components: The BioModel and the MathModel. The BioModel contains a description of the chemical and physical properties of the kind of cell the user is studying. Users then put an application to work in the model which defines the 3D geometry of the cell and the conditions for the model.
The MathModel is a mathematical description of this application and the resulting processes modeled in the cell. The MathModel provides researchers with an opportunity to access the underlying math directly.
VCell helps scientists expedite the scientific method by allowing them to explore cell behavior virtually before taking it into the lab. They can try different manipulations of the model to see how well their ideas would work out in an experiment.
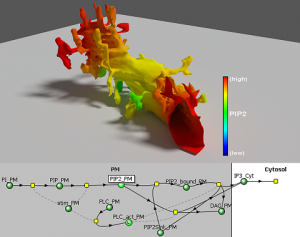
“You can’t really begin to form a hypothesis without help from computers to better understand these complex systems,” Loew says. “VCell allows you to do that even if you are not very well trained as a mathematician or physicist.”
Once researchers have created a model, they can publish it to the database and share it with the rest of the community on VCell alongside an academic paper of their findings. Other users can then reuse and manipulate the model for their own research, enriching the knowledge base and research potential of all users.
Over the years, VCell has grown its ability to analyze different kinds of problems of interest to researchers in different fields. VCell can now simulate about eight kinds of physiological problems in cells.
For example, VCell can simulate how the electrical action of a heart cell would change when a certain drug acts on those cells. These simulations provide scientists with valuable information about the effectiveness and possible side effects of drugs.
Loew says he hopes VCell will be able to expand its partnerships to include the pharmaceutical and biotechnology industries.
“We are constantly evolving to allow us to ask more and more different kinds of questions that require different simulation technology,” Loew says. “A primary focus for us over the next three years will be to nurture new partnerships with granting agencies, foundations and industry to assure that VCell continues to thrive and grow beyond the period of this grant.”
Ann Cowan, UConn Health professor of molecular biology and biophysics and deputy director of the Center for Cell Analysis and Modeling; Ion Moraru, UConn Health professor of cell biology; Boris Slepchenko, UConn Health associate professor of cell biology; and Michael Blinov, UConn Health assistant professor of genetics and genome sciences are all co-investigators for this grant.
Loew earned his Ph.D. in chemistry from Cornell University and completed his postdoctoral training at Harvard. He was the editor-in-chief of Biophysical Journal from 2012-2017. Loew’s research interests include developing and characterizing fluorescent probes of membrane potential and how the spatial organization of molecules in cells is used to control cell function.
This project is NIH Grant No.: 1 R24 GM134211-01.
Follow UConn Research on Twitter & LinkedIn.