University of Connecticut assistant professor of molecular and cell biology, Leighton Core hopes to answer new questions about RNA through a $2 million grant from the National Institute of General Medical Sciences.
Our bodies respond, on a cellular level, to environmental changes through a complicated process – turning on or off parts of our vast genome, expressing specific genes at specific levels to respond to specific situations.
However, experts are learning that this process may be even more complex than previously thought.
When our bodies need to produce proteins, necessary codes are found in our DNA. The process of transcription creates a messenger RNA that is an RNA copy of the DNA sequence. During translation, transport RNA then brings over the amino acids that correspond to the RNA code. These amino acids are joined together to create the protein. These proteins are responsible for everything that happens in our bodies from development, to immune responses and healing.
But there are some segments of our DNA that make RNAs that don’t code for any protein – so why do we have them?
This is the question Core hopes to answer.
Even though it is not translated into protein itself, noncoding RNA plays a critical role in regulating transcription of protein-coding genes. ncRNA can recruit gene activators or suppressors to control where, when, and to what degree proteins-coding RNAs are transcribed.
“These observations suggest that regulation of ncRNA biogenesis adds an intricate layer of control to overall gene expression levels,” Core says.
Currently, a significant obstacle to studying and understanding the role of ncRNA is being able to identify and correctly classify them.
Core’s project will develop new experimental and computational methods that can identify the transcripts of non-coding RNA that will also allow him to predict the function of these segments. He will then look to identify the mechanisms that regulate the production and destruction of ncRNA and determine the effect of ncRNA on the protein-coding gene transcription process of genes located near them on our genome.
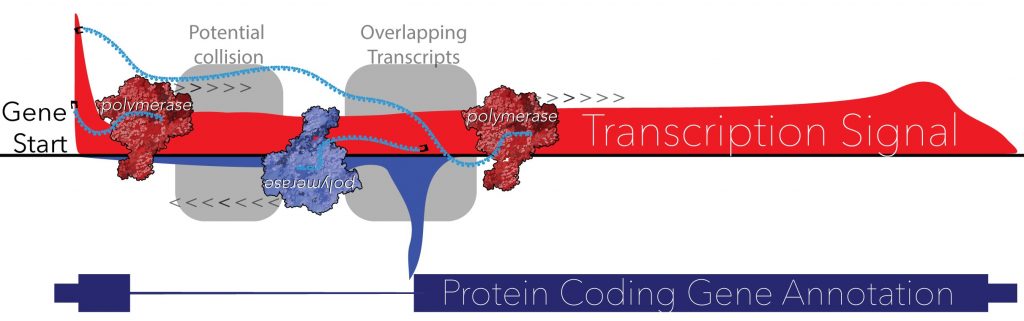
“ncRNAs are involved in regulating every phase of gene expression from transcription factor recruitment to RNA transcription, processing, stability, and translation,” Core says. “Intense efforts are still required to identify all species of ncRNAs and their diverse functions in regulating gene expression. These studies will generate important resources and represent foundational framework for studying ncRNA function.”
Core is also investigating the capacity of ncRNA in the evolution of new genes. He will use an evolutionary genetic model to identify the nucleotide changes in our DNA associated with altered ncRNA transcripts. This aspect of the project will shed light on the mechanisms underlying acquisition of new phenotypes and inform future studies of evolution.
Core received his PhD in molecular biology and genetics from Cornell University where he also completed his postdoctoral training. Core’s current research focuses on how changes in RNA transcription and processing drive changes or maintenance of cellular states during cellular responses such as development or disease progression.
This project is NIH Grant No.: Award #: 1R35GM128857-01



