Inflammation is part of the body’s natural response to injury or infection. It helps eliminate the cause of cell injury, clear away dead cells, and initiate tissue repair. While inflammation is generally beneficial, if left unchecked, it can damage key systems, including the cardiovascular system.
When the inflammatory response goes awry, it can contribute to vascular lesions. These lesions play a role in aneurysm, seizure, stroke, atherosclerosis, and other diseases.
UConn Health researchers Patrick Murphy, assistant professor in the Center for Vascular Biology, and Anthony “Tony” Vella, professor and chair of the Department of Immunology, have received a $2.7 million grant from the National Institutes of Health to study the role of RNA binding proteins and alternative splicing in regulating inflammation.
In the course of postdoctoral work at MIT, Murphy discovered the endothelial cells lining arteries responded to immune cell recruitment in the early stages of vascular injury by altering the composition of the mRNA through a process known as alternative splicing.
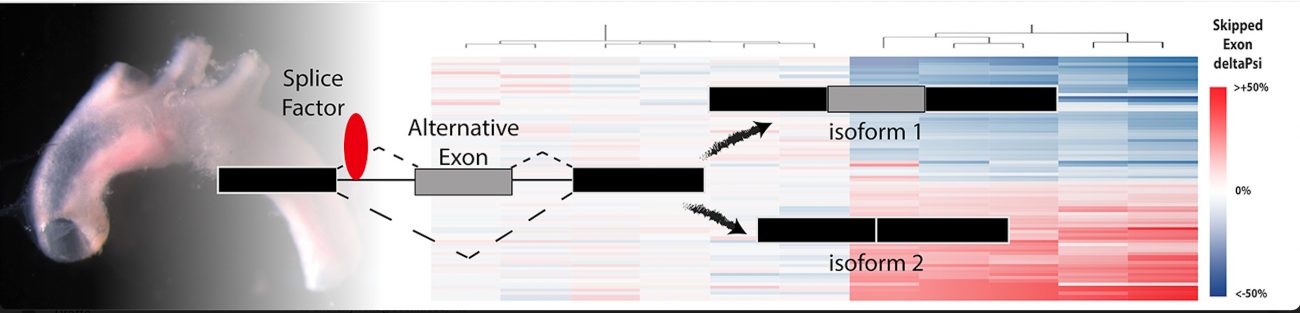
The pre-mRNA message derived from a single gene can be cut up many different ways and include different exons, or coding pieces of RNA. Depending on which exons are included in the final transcript, a different protein will be produced. Through alternative splicing a single gene can code for multiple proteins.
Endothelial cells are gatekeepers of immune infiltration into tissues, like the artery wall. Murphy found that blocking the alternative splicing of one gene increased the risk of arterial dissections and altered immune cell function. This led Murphy to hypothesize that alternative splicing regulates the nature of the interaction between the immune system and the arterial wall.
Supported by funds from the National Institutes of Health, the American Heart Association, and startup funds from the UConn School of Medicine, Murphy established in vitro screening approaches and in vivo models to attack this question in his new lab.
A talented team, led by biomedical sciences graduate students Jessica Hensel and Sarah-Anne Nicholas, and research assistant Amy Kimble, used CRISPR-KO screens to identify a set of RNA-binding proteins – splice factors – regulating these splicing responses and, unexpectedly, the inflammatory activation of endothelial cells.
Surprisingly, many of these RNA-binding proteins also bound transposon sequence in introns, areas of pre-mRNA transcript normally spliced out to make the final mRNA message. Transposons are self-replicating pieces of DNA that hop from one location in the genome to another. Although few transposons are active now, the genome is littered with their remnants, making up approximately 50% of our genomes.
Murphy thinks that by determining how much of this transposon-derived sequence is incorporated into an mRNA message, this small set of splice factors has driven the evolution of our inflammatory responses.
The key question then was: do transposon-binding splice factors modulate inflammatory responses in vivo? This is where Murphy received critical help from Vella.
“In my faculty search, I’d targeted research environments with strength in immunology,” Murphy says. “I’m happy to say that I got this one right. This work would not have been possible with the insights and help of the Immuno-Cardiovascular Group, and Tony Vella in particular.”
The Immuno-Cardiovascular Group, organized by Dr. Annabelle Rodriquez-Oquendo and led by Vella and Beiyan Zhou, has attracted top investigators in the cardiovascular field, including Zhichao Fan and Alison Kohan. They are applying their collective expertise and skills to attack a fundamental problem in cardiovascular disease – that most of the people suffering from heart attack or stroke already have well-controlled lipid levels.
Atherosclerotic plaques that cause heart attack and stroke occur through a buildup of lipids, fatty material in the artery wall. While lipid-lowering agents, like statins, have made significant strides in reducing the risks of these lesions, vascular “scars” remain even after bringing down lipid levels. Heart attack and thrombotic stroke are the major causes of death in the United States.
Encouragingly, recent work has shown these lesions can be targeted by directly modulating inflammation, indicating this is likely to be the next frontier in treating them.
The small set of transposon-binding splice factors Murphy identified are correlated with disease risk in human coronary arteries.
Murphy is working with Vella to test the hypothesis that transposon-binding splice factors play an important role in determining the outcome of inflammatory responses in the plaque. They will apply cutting-edge single-cell analysis tools to examine atherosclerotic plaque in animal models with inactivation of these genes – techniques highlighted in their recently published work.
Their work will help researchers and clinicians better understand why some atherosclerotic lesions can slowly smolder for decades, while other plaque becomes unstably inflamed, leading to heart attack and stroke. Ultimately, this knowledge could be used to identify at-risk patients as well as novel therapeutic approaches.
Patrick Murphy holds a Ph.D. in biomedical sciences from the University of California, San Francisco. He completed postdoctoral training at the Massachusetts Institute of Technology. His lab studies the interactions between recruited immune cells and the arterial walls using inducible genetic models.
Tony Vella completed postdoctoral training at the National Jewish Center for Immunology and Respiratory Medicine. His lab studies T cell and inflammation biology focusing on vaccine adjuvants, T cell costimulation, and pediatric intestinal inflammatory diseases.