Monkeyflowers glow in a rich assortment of colors, from yellow to pink to deep red-orange. But about 5 million years ago, some of them lost their yellow. In the Feb. 10 issue of Science, UConn botanists explain what happened genetically to jettison the yellow pigment, and the implications for the evolution of species.

Monkeyflowers are famous for growing in harsh, mineral-rich soils where other plants can’t. They are also famously diverse in shape and color. And monkeyflowers provide a textbook example of how a single-gene change can make a new species. In this case, a monkeyflower species lost the yellow pigments in the petals but gained pink about 5 million years ago, attracting bees for pollination. Later, a descendent species accumulated mutations in a gene called YUP that recovered the yellow pigments and led to production of red flowers. The species stopped attracting bees. Instead, hummingbirds pollinated it, isolating the red flowers genetically and creating a new species.
UConn botanist Yaowu Yuan, an associate professor in the Department of Ecology and Evolutionary Biology, and postdoctoral researcher Mei Liang (currently a professor at South China Agricultural University), with collaborators from four other institutes, have now shown exactly which gene it is that changed to prevent monkeyflowers from making yellow. Their research adds weight to a theory that new genes create diversity of appearance and even new species.
The YUP gene in question is found at a locus, or region, of the monkeyflower genome that has three new genes. These new genes are not found in species outside of this group. They are duplicates of other genes from other parts of the monkeyflower genome. In particular, YUP is a partial duplicate of a pre-existing gene that has nothing to do with color.
Standard genetics thought is that partial duplicate genes regulate the genes they are derived from; it was very unlikely that these genes would affect an unrelated gene. Liang decided to investigate what these genes were doing anyway, against the advice of Yuan, who thought it was a waste of time. But Liang’s persistence paid off: she discovered that the YUP gene was actually targeting the plants’ master regulator of carotenoids, the pigments that make monkeyflowers and other plants yellow. YUP produced many small RNAs that suppressed the carotenoid gene. There are very few examples of genes that produce small RNAs affecting traits important to the creation of a new species.
“This experience really taught me how important it is not to constrain oneself with ‘conventional wisdom,’” Yuan says. Not only does YUP regulate a gene it is entirely unrelated to it; the other two genes at this same locus also affect monkeyflower color, Yuan says.
The uniqueness of these three genes, only found in a few closely related monkeyflowers, is an important clue as to how new species evolve.
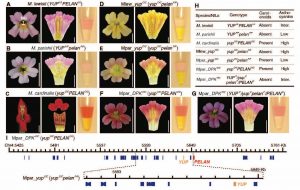
“Almost every single species has unique genes,” called ‘taxon specific’ because they are only found in a small group of species. “For the most part, we have no idea what these genes do,” says Yuan. This research shows that these taxon specific genes can be the keys to the new species. Previously, many geneticists and evolutionary biologists thought that it was changes in the expression of common genes shared by many different species that differentiated them, and that the small number of idiosyncratic genes were unlikely to be important.
“We think we understand evolution well enough to make predictions. But now we are realizing we really don’t. Evolution is just so unpredictable,” Yuan says.
His lab is now looking at how the monkeyflower genome controls the production of pigment spatially. For example, some monkeyflowers have upper petals that are entirely white, but lower petals with color. Yuan and his colleagues want to know how the plants suppress pigment only in certain parts of the flower.



