It may seem surprising that relatively little is known about the genetics of many of the microscopic animals at the base of the marine food web. The reason for this is that until now it has not been possible to study their diverse genomes for the most part or, in strikingly many cases, even just to culture them in the laboratory. An international group of scientists from over 40 research groups, including UConn marine science professor Senjie Lin’s team, collaborated to create the tools and protocols necessary for studying the genetic details of a broad range of these microscopic marine creatures called protists. Their work was published today in Nature Methods.
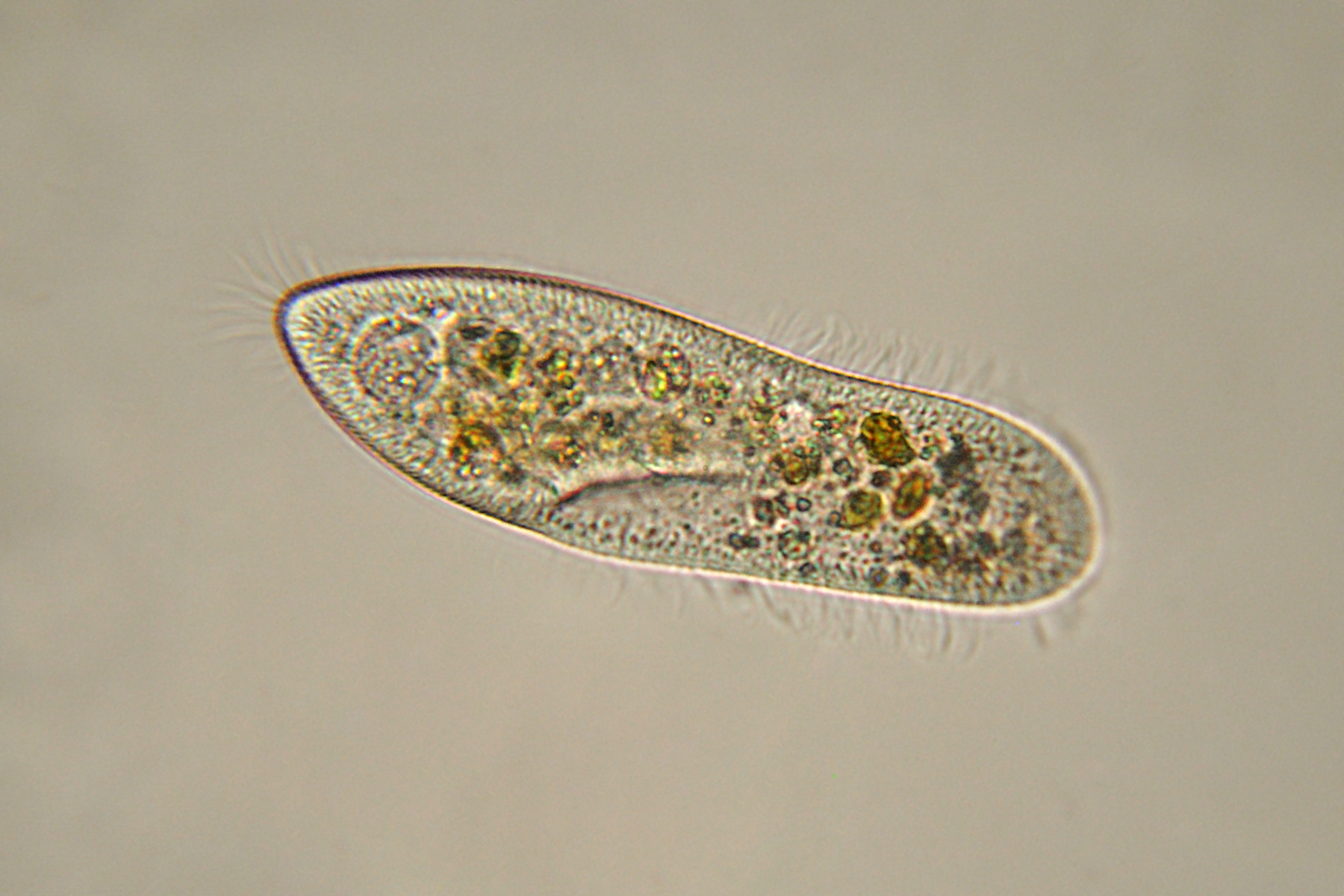
Lin says protists occupy the largest part of the tree of life and play key roles in the marine environment, from being important components in the food web to cycling elements within marine environments. However, even when looking at only one group, such as dinoflagellates, one would be amazed how diverse their lifestyles are.
“Some species are photoautotrophic, utilizing sun light and carbon dioxide as an energy source and cell backbone,” he says. “Some are heterotrophic, growing on available food; yet some others are mixotrophic, which can be both photoautotrophic and heterotrophic depending on what is available. Some dinoflagellates are parasitic, able to infect fish, crabs and other crustaceans, and even their own dinoflagellate fellows. Some other dinoflagellates are important symbionts to treasured coral reefs. Yet, very little is known what genetic makeup underlie these different life styles.”
To study these details in an organism, it is extremely helpful to be able to experimentally manipulate its genome. This requires researchers to culture the organisms, extract the DNA, and then insert genes into the genome to learn more about the specific gene functions. Though the process, called transformation, may sound relatively straightforward, developing protocols for genetic engineering can be quite complex for a single species, let alone for a diverse range of species.
Lin says his project was funded by the Gordon and Betty Moore Foundation under the Marine Microbial Initiative program. In 2012, the Foundation sponsored a Marine Microbial Eukaryote Transcriptome Sequencing Project to fund sequencing of several hundred species or strains of protists.
“My lab participated in that project and had several dinoflagellate species sequenced,” says Lin. “The gene transformation project is a natural extension of the Moore Foundation’s endeavor to drive forward biology of marine protists, with the objective of enhancing the understanding of how marine protists respond and adapt to the diverse marine environments and play roles in nutrient cycling.”
The researchers approached the task by utilizing data from the genome sequencing initiative. Then they systematically analyzed genes to identify and optimize steps that would result in effective transformation protocols. They called the resulting information the synthetic “Transformation Roadmap” that helped identify strategies to apply throughout the transformation process, from ideal culture conditions to which method of gene insertion may be most successful for a particular species.
Brittany Sprecher, a PhD student in Lin’s lab, says the work was fun and yet frustrating at times. “In our lab we started with at least seven different dinoflagellate species and were able to successfully carry out transformation with just two species after several years,” she says. “These two successes required a lot of trial and error in every step of the process, adapting protocols for our unique dinoflagellates. We were very fortunate to have this community to help bounce ideas off of during our frequent virtual conferences.”
All in all, this collaboration has taken the number of species that had effective, reproducible transformation protocols from only a few protist species to 13 species that had never been successfully transformed before. The researchers were also able to make progress towards successful transformation in an additional eight species, and elucidated possible reasons for why 17 other species were not transformed successfully.
Lin says, “It is fantastic that we can now study 13 more protists in great detail with these protocols and can add to the list of ‘model organisms.’ One of the greatest treasures of this paper and community coming together was to share what was attempted with the 17 other species that were not able to be transformed. We wanted to make sure to share the different trials and errors for the future experimenter to see and build off of. As many people in science know, it is great to see studies that didn’t work as well as the ones that worked.”
Associate research professor and member of Lin’s team Huan Zhang says, “Being a member of this collaborative network has been such a blessing and emphasizes the need for more research collaboration. It was also great that the Moore Foundation understood the challenges and difficulties of these projects and worked to bring us together constantly”.